A new tool can make any type of edit—large or small—to RNA, vastly expanding the scope of the approach overall, researchers say. More adaptable and efficient than its predecessors, the technique is aptly dubbed Programmable RNA Editing & Cleavage for Insertion, Substitution, and Erasure—or PRECISE, for short.
PRECISE enables “nuanced and precise genetic manipulation,” co-lead investigator Omar Abudayyeh, a fellow at the Massachusetts of Technology (MIT), wrote in an email to The Transmitter. “Researchers can now investigate the effects of various edits, including those that were previously challenging or impossible to achieve, such as large deletions or insertions, facilitating deeper insights into gene regulation, expression, and the impact of mutations on cellular functions.”
DNA editors such as CRISPR and prime editing are physically large and therefore difficult to deliver to cells. They also run the risk of permanent off-target effects.
By contrast, with RNA editing, “there is no long-term alteration of the genome made at the level of DNA,” says Ethan Bier, professor of cell and developmental biology at the University of California, San Diego, who did not take part in the new research. And RNA editors are typically simpler than DNA editors and easier to deliver to a wide range of tissues.
Until now, RNA editing approaches were limited to swapping only two sets of 12 possible base replacements. Nor could they insert or delete new bases.
But PRECISE is capable of making all of the base replacements, as well as insertions ranging from 1 to 1,863 base pairs and deletions of up to 24 base pairs. Its creators contend it can make insertions or deletions of any size and can target about 96 percent of the 141,342 known pathogenic mutations in the human genome.
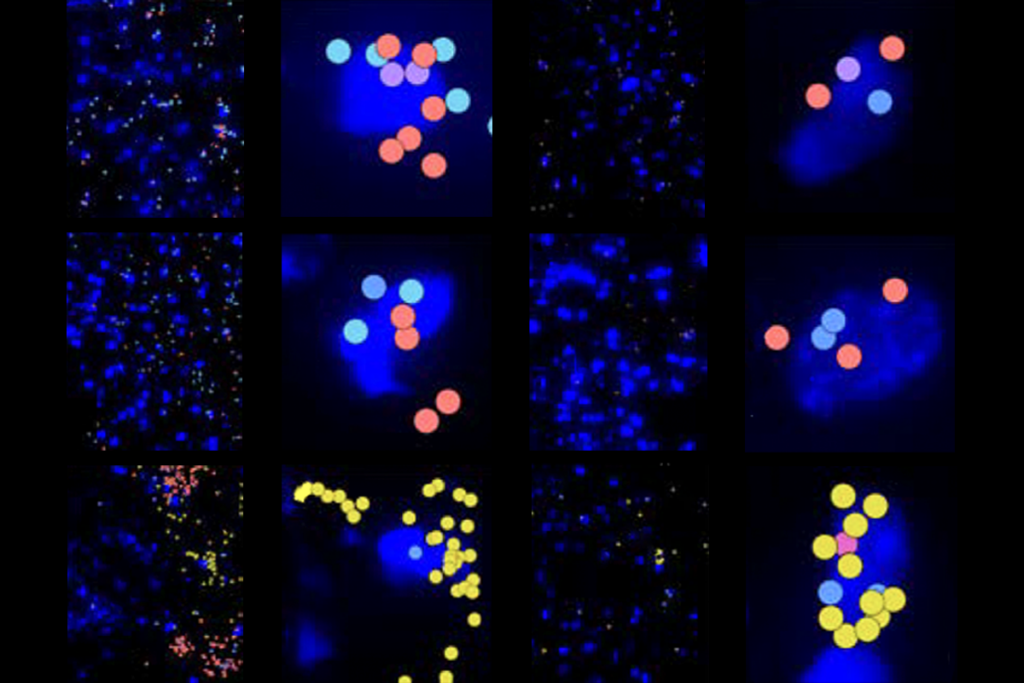
For example, PRECISE reached up to 30 percent efficiency when editing RNA transcripts of the SHANK3 gene, which is linked to autism, in human embryonic kidney cells, among other proof-of-concept experiments described in a preprint posted on bioRxiv in February.
The technique might also be used to create new models and treatments for autism, says co-lead investigator Jonathan Gootenberg, a fellow at MIT.
P
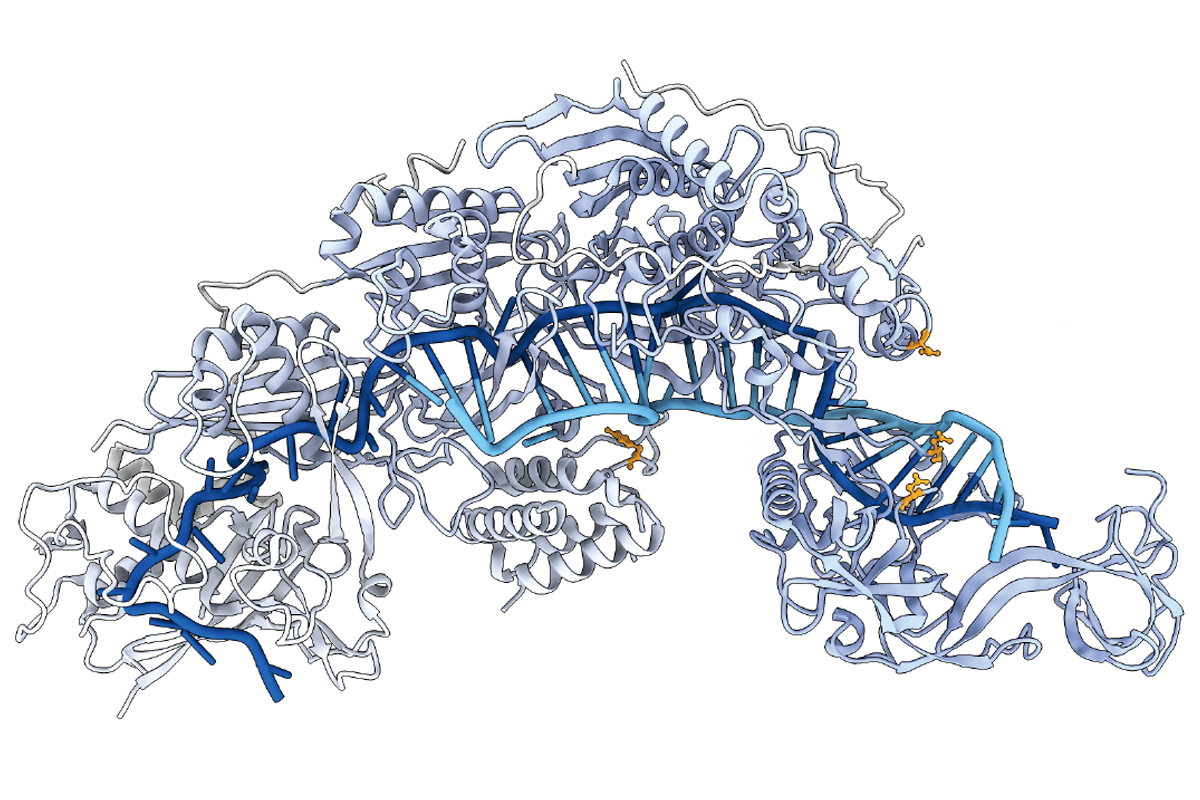
RECISE modifies the output of gene transcription by way of trans-splicing, or joining two RNA molecules together, and can modify bases starting from either the 5’ or 3’ end of an RNA strand, using programmable RNAse proteins.PRECISE modifies 5 to 50 percent of transcripts when applied to nine different kinds of cellular RNAs. That efficiency rate is 10 to 100 times better than previous trans-splicing RNA editors. It owes part of its success to the recently discovered CRISPR-Cas7-11 bacterial enzyme, which can make specific cuts in target RNAs, easing the way for trans-splicing to occur.
The new technology is “extremely interesting,” says Joshua Rosenthal, senior scientist at the Marine Biological Laboratory in Woods Hole, Massachusetts, who was not involved in the work. “If this technology can really be harnessed efficiently, it could be ground-breaking.”
The downside of using bacterial proteins is that they might trigger immune reactions, says Andrew Lever, professor of infectious diseases at the University of Cambridge, who was not involved in this study. And because an RNA editor’s effects are not permanent, they would need to be delivered repeatedly for lasting effects, potentially increasing the chances of immune problems, he adds.
In an effort to avoid such problems, for 5’ trans-splicing, the scientists behind PRECISE developed an additional mechanism that employs a catalytic RNA molecule known as a ribozyme instead of a bacterial protein. This approach is simple enough to deliver a payload about 3,700 bases long in a single adeno-associated virus (AAV) and achieved up to 15 percent editing efficiency in human embryonic kidney cells.
“The protein-free version of PRECISE editing is incredibly easy to deliver, using single-vector viral delivery, unlike more complex DNA editors, like base editors and prime editing,” Gootenberg says.
This version also achieved 4.5 percent editing efficiency in targeting the Huntington’s-related gene HTT in human cortical neurons. And PRECISE appeared to cause no significant changes to normal expression levels of other genes or any other off-target effects.
Using RNA editing for conditions such as autism may prove “extremely challenging,” Rosenthal says, because it would likely need to target the developing brains of young children to work.
But, Abudayyeh notes, “AAV delivery is rapidly improving and will hopefully one day be ready to use in younger and younger patients as safety and efficacy is demonstrated in a responsible and ethical way.”