‘Retro-Cascorder’ tracks gene-expression timing
The new tool may help researchers reconstruct the sequence of biological events that underlie development.
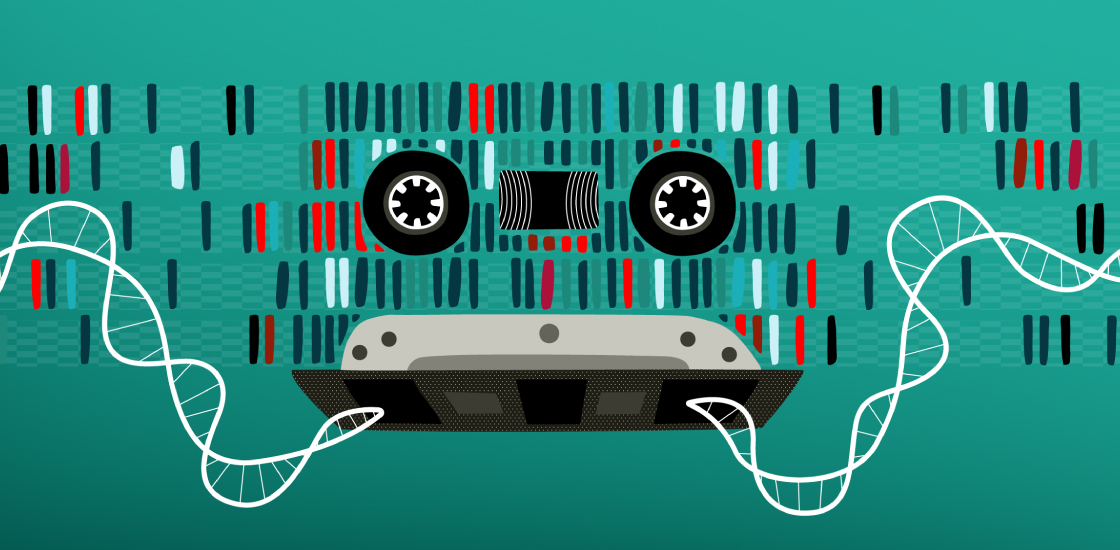
A new molecular system records the expression of different genes over time in a cell’s DNA, making it possible to reconstruct the order of biological events within individual cells. The tool could expose the processes that underlie cellular differentiation during development and how they may be altered in autism.
Researchers typically track gene expression by sequencing RNA, the intermediary between genes and proteins, in batches of cells at consecutive time points. But biological events are not perfectly synchronized across different cells, making it difficult to reconstruct processes precisely.
Another approach involves registering cellular processes in a portion of a cell’s genome. Devices called ‘molecular recorders’ can document the occurrence of a particular biological event, such as a cell’s response to a specific molecule, in DNA. They can also log arbitrary, time-ordered information into the genome: In 2017, Seth Shipman, assistant investigator at the Gladstone Institutes in San Francisco, California, and his colleagues encoded a short film into the DNA of bacteria.
To gain insight on neuronal differentiation and brain development, however, researchers need to be able to capture the order of a cell’s internal events. “That, traditionally, has been a really hard thing to do,” says Shipman, who led the new work.
The novel tool Shipman and his colleagues developed, called a Retro-Cascorder, converts RNA barcodes into DNA and integrates them into a section of a cell’s genome in a unidirectional manner, creating a chronological recording of its gene expression. By sequencing the cells, researchers can then assess the position of the relative barcodes to deduce the order in which genes were activated, the researchers reported in July in Nature.
T
o create the Retro-Cascorder, Shipman and his colleagues combined two components of the bacterial immune system: retrons and part of the CRISPR system.Retrons typically encode an RNA sequence along with an enzyme that generates a DNA copy of it. The team modified a six-letter segment of a retron to create nine distinct RNA barcodes, which are converted into DNA. By engineering cells to co-express one of the barcoded retrons when a specific gene is turned on, scientists can generate DNA tags of gene expression, the researchers reported.
To record these DNA tags, the researchers turned to two CRISPR enzymes — Cas1 and Cas2 — that can add bits of foreign DNA to one end of a section of the bacterial genome called a CRISPR array. The array archives the DNA tags expressed in a cell over time, with those located farther along the array having been generated earlier in the recording.
Overexpressing retrons along with Cas1 and Cas2 in cells of Escherichia coli bacteria revealed that retron-derived DNA segments are integrated into CRISPR arrays. And DNA sequencing accurately distinguished six RNA barcodes that differed by at least four letters.
The researchers then tested the system’s ability to track gene expression over time. They introduced a pair of plasmids, or DNA loops, into E. coli. One expressed Cas1, Cas2 and the enzyme that converts retron RNA into DNA. The other carried two genes tagged with distinct barcoded retrons whose expression is activated by different chemicals. They exposed cells to each chemical for 24 hours, reversing the order of exposures in one batch of cells.
By analyzing CRISPR array sequences after 48 hours, the researchers were able to accurately determine the order of both courses of chemical exposures, they reported.
Sequences that were not derived from retrons also accumulated in the CRISPR arrays throughout the experiment, which jibes with previous research that suggests that Cas1 and Cas2 recognize free-floating bits of DNA in cells. These random sequences are added into the array at a relatively constant rate, so they may be useful for gauging how early or late in a recording a retron tag is expressed, the researchers say.
The retron-based approach seems to work well, according to Junhong Choi, a postdoctoral fellow in Jay Shendure’s lab at the University of Washington in Seattle, who was not involved in the work but recently developed another tool that records cellular events into DNA, which was also described in a paper in Nature in July. “I haven’t seen anything like that before,” Choi says. Currently, however, systems that rely on Cas1 and Cas2 have been used only in bacteria, he says.
According to Shipman, the tool needs to be tweaked before it can be used in neurons. He and his colleagues are trying to optimize the system and transition it to eukaryotic cells. “We’re making progress,” he says, “but there is plenty of work to be done.”
Recommended reading
Brain ‘chimeroids’ reveal person-to-person differences rooted in genetics
Widely used calcium imaging protocol can lead to spurious results, new paper cautions
Robots boost data consistency in rodent studies reliant on mechanical, optogenetic stimulation
Explore more from The Transmitter

Building an autism research registry: Q&A with Tony Charman