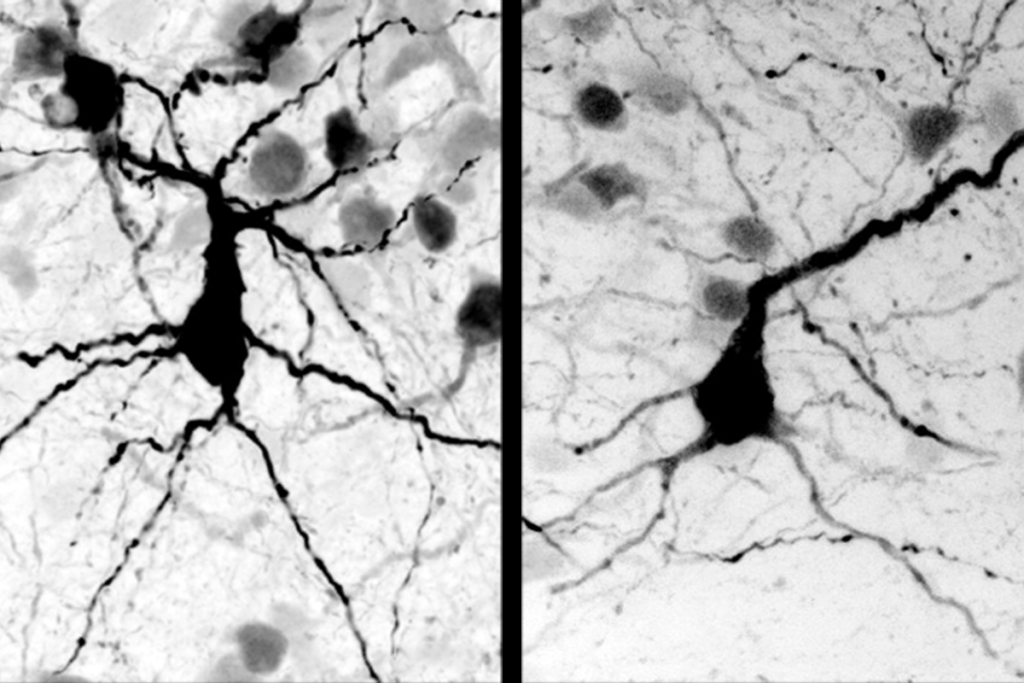
Measuring expression
To examine protein interactions inside brain cells, scientists typically zero in on one gene at a time. A new method described in today’s Nature simultaneously measures expression of the whole genome.
Brain cells are in constant motion. Ions rush in and out of synapses, the nubs that transmit messages between neurons, and trigger hundreds of protein interactions. Somehow, this chain of signals travels down neuronal branches to the cell body, where it can affect the expression of thousands of genes.
To study these complex networks, scientists typically zero in on one gene — or even a part of a gene — at a time, then gradually work backwards to figure out what turned it on, when and why.
That targeted approach seems ridiculously tedious compared with a new method that gives scientists the ability to simultaneously measure expression of the entire genome. A study applying the technique in brain cells is described today in Nature.
I was spellbound when neurobiologist Mike Greenberg presented a preview of this rigorous work in a keynote lecture Sunday evening at the Keystone symposia on autism in Snowbird, Utah.
Using a technique called RNA-Seq — which decodes super-tiny bits of messenger RNA, the molecules that help translate DNA into protein — Greenberg’s team at Harvard compared whole-genome expression patterns of a neuron before and after artificially stimulating it in culture.
Focusing on the genomic region surrounding c-FOS, a cancer gene whose expression is influenced by many signals from outside the cell, they discovered that c-FOS expression increases dramatically after the cell is stimulated. In parallel experiments, they found that neuronal activity also drives the production of strings of enhancer RNA, a new class of RNA that binds proteins to up the expression of distant genes.
Taken together, the findings suggest that, once a neuron is stimulated, enhancer RNA somehow hops across large distances to turn on c-FOS. Traditional experiments, which look mainly at promoter sequences right next to a gene, would never have uncovered this piece of the puzzle.
In his talk, Greenberg said what’s most striking about this network of interactions that stretches from a stimulated synapse to the nucleus is that it’s “studded with proteins” that are mutated in cognitive disorders.
For example, MeCP2, the gene mutated in Rett syndrome, silences other genes. Children carrying the mutation are fairly normal until about 1 year of age, when they undergo a dramatic regression and, often, seizures. Tellingly, 1 year is also when many synapses are being shaped by experience.
What I find exciting is that this is just the beginning. Because they took an unbiased whole-genome approach, Greenberg’s team is left with much, much more data than they know how to interpret — at least for now.
Recommended reading
New organoid atlas unveils four neurodevelopmental signatures
Explore more from The Transmitter
Snoozing dragons stir up ancient evidence of sleep’s dual nature

The Transmitter’s most-read neuroscience book excerpts of 2025