New sequencing technique may reveal recessive mutations
A new method of genetic analysis allows researchers to identify regions that are identical on both copies of a chromosome, according to a study published 20 September in Molecular Cytogenetics.
A new method of genetic analysis allows researchers to identify regions that are identical on both copies of a chromosome, according to a study published 20 September in Molecular Cytogenetics1.
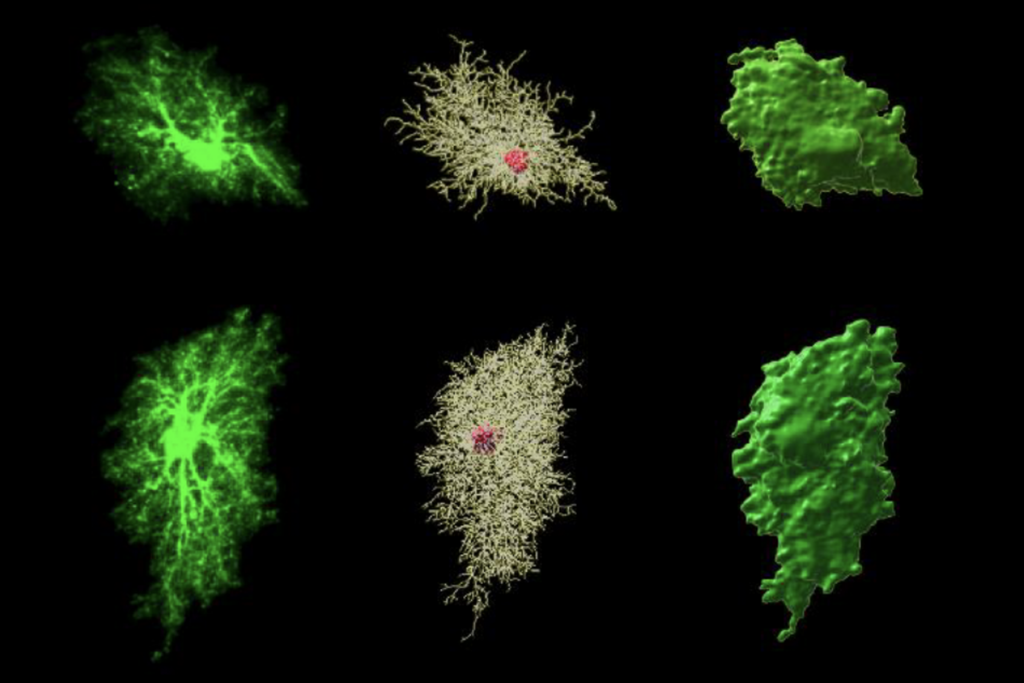
People who have autism are more likely to have these so-called ‘regions of homozygosity’ than their unaffected siblings. Scouring these regions has helped reveal new autism candidate genes.
The genetics of autism is a complex combination of rare, harmful mutations and common variants. Recessive mutations reveal their effects only when inherited from both parents. For this reason, researchers often look for mutations linked to autism in populations in which cousin marriages, which increase the likelihood of two parents sharing genetic information, are common.
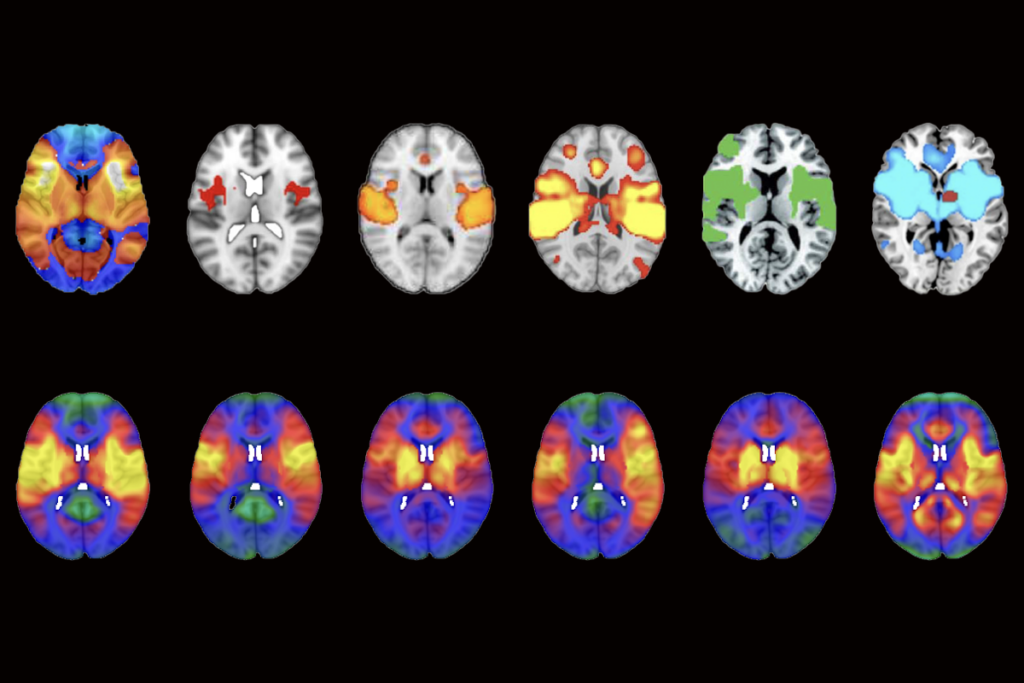
In the new study, the researchers developed a method to find regions of homozygosity in the genome. The method makes use of genomic microarrays, which match pieces of an individual’s DNA with snippets of reference DNA, or probes. The new method uses probes designed to detect duplications or deletions of DNA as well as more subtle variations in single DNA nucleotides.
The researchers tested 607 children for homozygosity: 21 of these children are likely to have closely related parents, they found. The researchers were able to follow up on parental relationship for 14 of the 21 children. The parents were found to be unrelated in only one case. The relationships range from brother and sister to being from the same town.
The new method, which reliably identifies potential sites of recessive mutations, may help researchers home in on autism candidate genes.
References:
1: Fan Y.S. et al. Mol. Cytogenet. 6, 38 (2013) PubMed
Recommended reading

Common and rare variants shape distinct genetic architecture of autism in African Americans
Profiles of neurodevelopmental conditions; and more
Explore more from The Transmitter
How artificial agents can help us understand social recognition