Atlas maps ‘junk’ DNA that may regulate brain development
Researchers have cataloged thousands of DNA regions that may act as enhancers — regulating gene expression from afar — in the developing mouse and human brains, according to a study published 14 February in Cell.

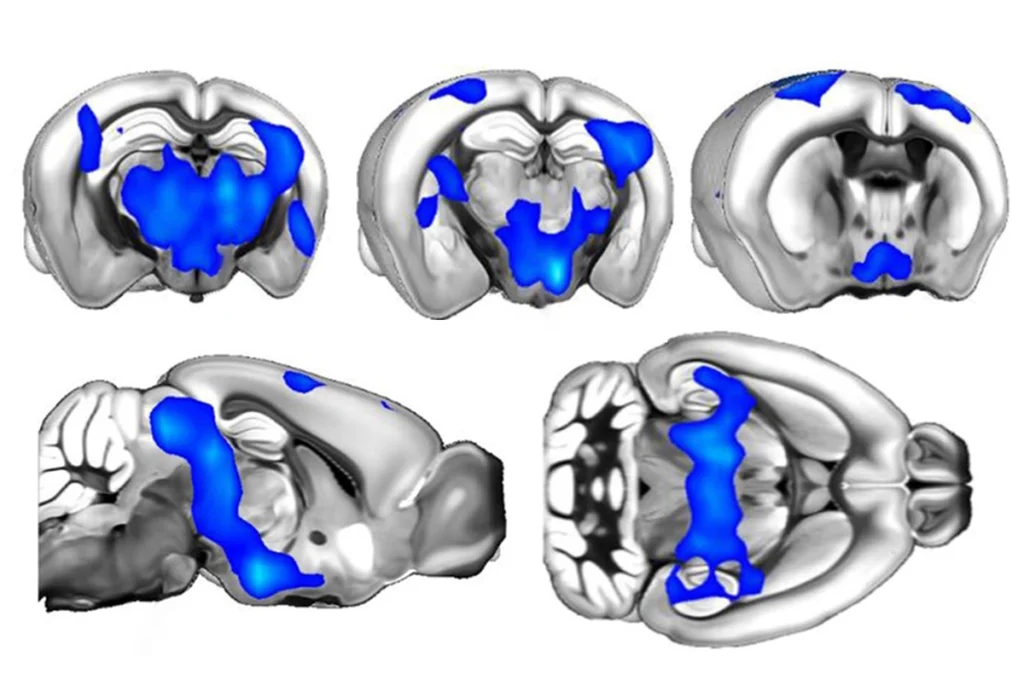
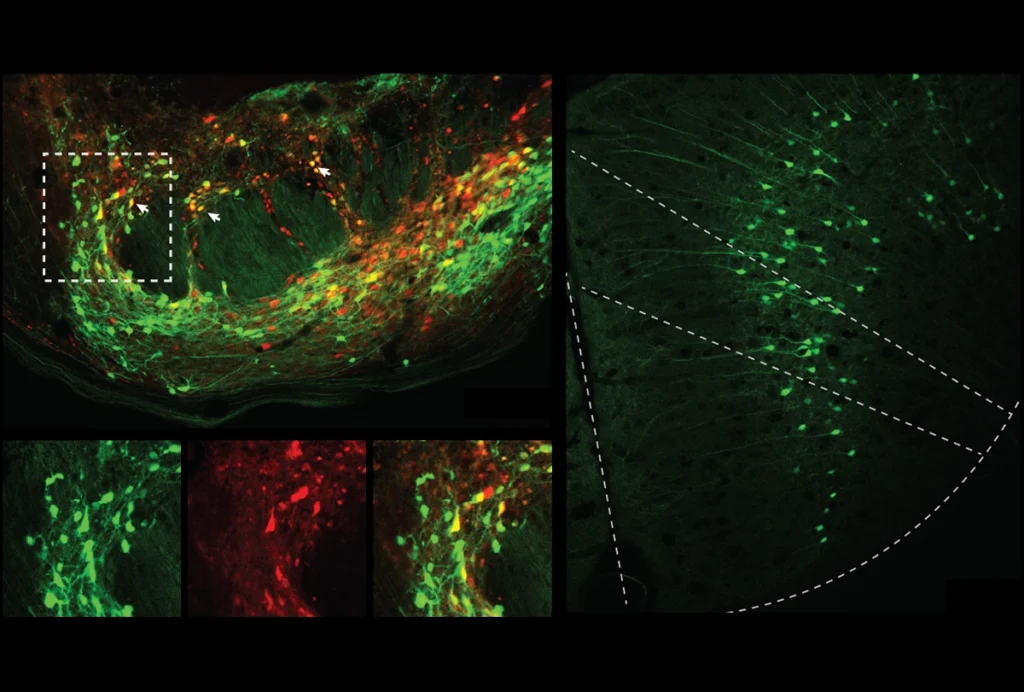
Brain drivers: Enhancer sequences turn on gene expression at specific regions in the developing mouse brain.
Researchers have cataloged thousands of DNA regions that may act as enhancers — regulating gene expression from afar — in the developing mouse and human brains, according to a study published 14 February in Cell1.
The report also paints a detailed picture of the mouse brain sites at which many enhancers activate their targets.
Several studies have mapped patterns of gene expression in the brain, showing where and when certain genes become active. But less well known are the DNA elements that may regulate these patterns, especially those that are physically removed from their genetic targets.
Because these enhancers do not code for protein, their importance is not obvious. An enhancer catalog will allow researchers to prioritize disease-linked variants in DNA that does not code for protein. They can also use this so-called ‘junk DNA’ to drive gene expression in certain brain regions at particular periods in development.
Using an enhancer-binding protein, the researchers identified 4,425 potential enhancers in the developing mouse brain. They also found 231 sequences that are near genes important in brain development and that are highly evolutionarily conserved.
Using the same technique, they also identified 2,275 potential enhancers in the cortex of a 20-week-old human fetus. Of these, 58 percent match candidate enhancers from developmentally equivalent mice; 421 are present in duplications or deletions of DNA that are more common in people with autism than in controls.
Of 329 potential enhancers chosen at random from among the mouse candidates, 105 activate expression of a blue reporter protein in embryonic mouse brains. The researchers used high-resolution microscopy to map where the enhancers drive expression of this reporter, and presumably their gene target, in the brain. Images of 32,000 sections of the developing mouse brain are available online.
The researchers also linked an enhancer region to CRE, an enzyme used experimentally to switch gene expression on or off. Using this technique, researchers can manipulate gene expression in certain regions of the brain during development.
References:
1. Visel A. et al. Cell 152, 895-908 (2013) PubMed
Recommended reading

Building an autism research registry: Q&A with Tony Charman
CNTNAP2 variants; trait trajectories; sensory reactivity
Brain organoid size matches intensity of social problems in autistic people
Explore more from The Transmitter
Cerebellar circuit may convert expected pain relief into real thing