‘Master switch’ molecule keeps overexcited brain in check
Some small fragments of RNA are expressed differently in people with autism than in controls, according to two new studies. The findings unveil another layer of complexity in the genetics of autism.
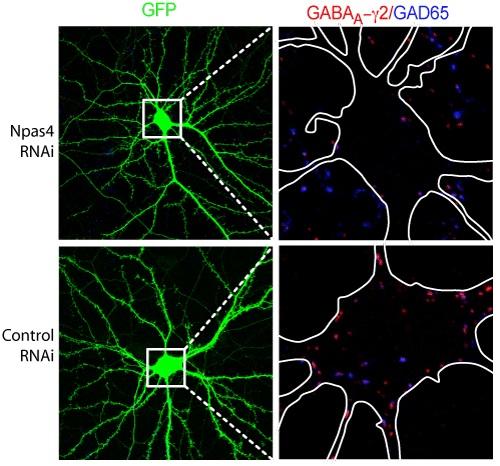
Damped down: There are far fewer GABAergic synapses in cultured mouse hippocampal neurons in which Npas4 is knocked down by RNAi (top right) compared with control neurons (bottom right).
Researchers have uncovered a new molecular mechanism underlying the balance between excitatory and inhibitory neurons in the brain ― a balance that can go awry in neurological disorders such as epilepsy, autism and schizophrenia.
Experts say the findings1, published in Nature, are a first step toward helping unlock new molecular targets for treatment of these disorders.
In a series of tests on cultured rat and mouse neurons, the group assessed gene activation in neurons before and after exciting them using calcium to create a positive electrical charge inside their cell membranes.
The researchers found that a transcription factor called NPAS4 is activated in excitatory neurons after depolarization. NPAS4 in turn activates or represses about 270 genes, which help control inhibitory neuronal connections and may keep over-excitation in check, the researchers say.
The researchers also created mice that lack the NPAS4 gene. These mutant mice are anxious, hyperactive and prone to seizures, compared with controls. They also have consistently lower levels of brain-derived neurotrophic growth factor, a molecule previously implicated in Rett syndrome and other neurological disorders.
“The function of this protein [NPAS4] has not previously been characterized in any significant way,” says lead investigator Michael Greenberg, director of neurobiology at Children’s Hospital Boston.
In 2004, based on molecular and genetic analyses of NPAS4 in mice, researchers proposed that the protein is a transcription factor involved in brain development and function2. Since then, others have shown NPAS4 to be activated in rodent brains following seizures and strokes3,4.
Greenberg and his colleague, Chris Walsh, linked NPAS4 to autism in July, when they found greater numbers of mutations in the NPAS4 gene and several other activity-dependent genes in people with autism, compared with healthy controls5.
“There’s good reason to think the Npas genes are involved in cognitive disorders,” says Gordon Fishell, professor of cell biology at New York University. “Based on the work in this study and their collaborative effort with the Walsh laboratory, [NPAS4] smells right to be an important clinical target.”
Checks and balances:
Exactly how NPAS4 is related to developmental disorders such as autism is unclear, however.
Previous studies have shown that other autism candidate genes, such as neuroligin and neurexin, are also involved in maintaining a balance between excitatory and inhibitory connections in the brain6. Whatʼs more, mutations in genes that control the maturation of inhibitory interneurons ― which dampen electrical signaling in the brain ― trigger increased anxiety and spontaneous seizures in mice7.
During early brain development, excitatory neurons dominate the brain. Inhibitory neurons, which signal using the neurotransmitter Gamma-aminobutyric acid or GABA, sprout up in early childhood and adolescence to shape the networks and prevent over-excitation.
Studying exactly how NPAS4 is activated will help researchers understand how the balance between inhibition and excitation is established, says Steven McKnight, a professor of biochemistry at the University of Texas Southwestern Medical Center.
“Just finding out what this transcription factor does is a big step forward in determining how the brain gets wired with inhibitory neurons,” McKnight adds.
Greenbergʼs group plans to study the genes that NPAS4 targets, as well as how neuronal connections develop in mice that lack NPAS4.
But studying the slew of NPAS4 targets is likely to be a challenge. Of the 270 targets identified, 94 are uncharacterized; the characterized targets have diverse functions, including molecule trafficking and receptor endocytosis, that could influence the formation of new synapses.
“It will be an ongoing slog to figure out the logic of all those genes,” McKnight says.
References:
Recommended reading
Okur-Chung neurodevelopmental syndrome; excess CSF; autistic girls

New catalog charts familial ties from autism to 90 other conditions
Explore more from The Transmitter

Karen Adolph explains how we develop our ability to move through the world
Microglia’s pruning function called into question