We found a major flaw in a scientific reagent used in thousands of neuroscience experiments — and we’re trying to fix it.
As part of that ambition, we launched a public-private partnership to systematically evaluate antibodies used to study neurological disease, and we plan to make all the data freely available.

The most common genetic cause of frontotemporal dementia is a mutation in the gene C9ORF72, first pinpointed in 2011. This amazing discovery was followed by years of confusion about the cell biology of the protein C9ORF72 encodes. Some papers reported that it was localized to the nucleus, others to a range of different organelles, still others to the cytoplasm. And without a basic understanding of where the protein acts, there was no way to move toward developing a treatment for frontotemporal dementia.
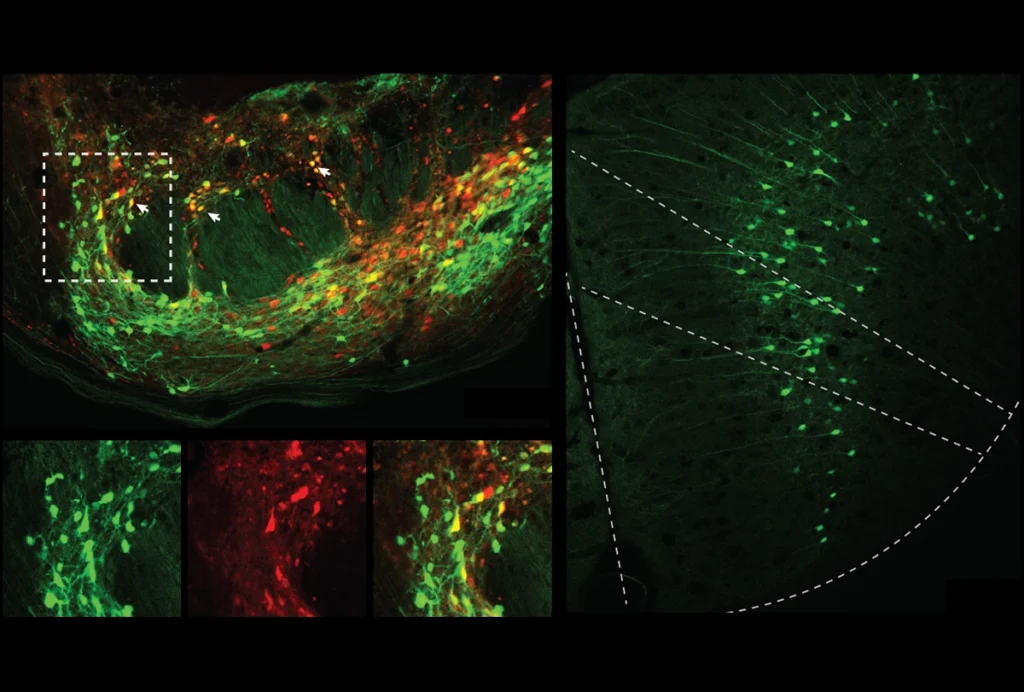
In 2019, working with a group of colleagues, we decided to look into the problem, focusing first on characterizing the antibody reagents other scientists had used to localize the protein. What we found was distressing. Using cells lacking C9ORF72 as controls, we discovered that not a single antibody reagent used in any of the published studies actually worked as advertised — they all bound to other proteins in addition to the target. In short, all the studies published using these C9ORF72 antibodies were potentially flawed. We also tested other commercial antibodies that had not, to our knowledge, been used in published research, and we found a few that were highly selective. These antibodies revealed that C9ORF72 is localized to the peri-lysosomal region and mostly expressed in microglial cells, discoveries that have since been replicated.
The C9ORF72 example illustrates a widespread problem. Antibodies are one of the most commonly employed reagents in molecular research, used to identify single proteins in a cell’s complex mixture. But scientists have known for decades that they can be inaccurate, binding to more than just the protein of interest. Publications that unknowingly use inaccurate antibodies can compound the issue, making it difficult to reproduce scientific results and raising questions about the validity of some preclinical drug studies.
Despite the seriousness of the problem, the field lacks a systematic way to characterize antibody accuracy. Quantifying how precisely an antibody highlights its target — its selectivity and specificity — is expensive and time consuming. The gold-standard approach is to compare cells expressing a target protein and those genetically modified to lack the target protein. Though many manufacturers do some knockout testing, the process is too expensive to apply to all antibody products. Most labs lack the requisite technologies, time or funding to rigorously characterize antibodies. As a result, most homemade or commercial antibodies are not subject to strict testing, a serious structural failure in the antibody ecosystem.
To address these structural issues, a team of us in academia and industry has launched an effort to systematically characterize widely used antibodies, employing knockout cells and tissues as controls whenever possible. Known as YCharOS, this public-private partnership has initially focused on antibodies used to study neurological conditions, including Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), autism, frontotemporal dementia and Parkinson’s disease. We plan to eventually characterize high-performing antibodies for all human proteins.
In a pilot project, we tested 614 antibodies to each of 65 proteins linked to ALS, Alzheimer’s disease and Parkinson’s disease, using knockout cell lines we generated or collected from academic and commercial partners. The results were sobering. Many commercial antibodies did not perform as expected — 60 percent of the antibodies were not specific to their intended target. Given this figure, we predict that as much as 20 percent of the figures in publications using these antibodies are in question. On the positive side, we estimate that well-performing antibodies are already available for more than 50 percent of protein targets. With this knowledge, antibody makers can focus their development efforts on the other 50 percent.
In just three years, YCharOS has already produced valuable information for the neuroscience community. But we could make faster progress with greater community contribution — particularly with community-supported sharing and generation of knockout cell lines.
A
ll the data we generate will be publicly available after internal scientific review. Indeed, the open-science nature of the project was essential in making it work — companies and others are willing to participate because the data are open. Companies in the partnership have donated more than 800 antibodies for our pilot project, for example.To date, YCharOS has published 78 antibody characterization reports for targets associated with ALS, Alzheimer’s disease, autism, frontotemporal dementia and Parkinson’s disease. These reports are available in the public domain at Zenodo, a general-purpose open-access data repository operated by the European Organization for Nuclear Research, CERN. To increase awareness, many of these reports have been shared via F1000 or more traditional publications, which enables them to be indexed on PubMed. Our protocols for immunoblotting, immunofluorescence and immunoprecipitation, developed in consultation with an expert group of academics and industry representatives, are also now publicly available. The protocol for immunohistochemistry is in development.
The reaction from commercial providers has been outstanding. When antibodies perform inadequately in the mutually agreed-upon standard operating procedures, their makers either remove the antibodies from the market (18 percent of antibodies tested) or alter their recommended usage (37 percent of the antibodies tested). We are also seeing an increase in the use of well-performing antibodies in the literature.
Manufacturers have also adjusted their research and development pipelines to meet the gaps that YCharOS has identified. For example, when it was clear that no existing antibodies were available for the proteins PRKN and SMOC1, two companies produced new well-performing antibodies. Similarly, based on feedback from YCharOS, many manufacturers are gearing up their pipelines to produce new recombinant antibodies to HTT, the protein implicated in Huntington’s disease.
Looking forward, the field needs to address the availability of knockout cells, a key bottleneck. We could assess more antibodies if individual scientists shared their lines with YCharOS and if funders supported concerted community efforts to generate knockout lines. Scientists interested in a specific target could also include a budget line item to support the characterization of all commercial renewable antibodies for that target in each of many applications — we estimate this would cost roughly $50,000 per target. All these efforts would have a lasting impact because the characterization is focused on renewable and recombinant antibodies.
We have a clear scientific road map to identifying well-performing antibodies for all human proteins and an organizational framework to carry out this task. If the community works together, we will get there faster.

University of Toronto

YCharOS and the Structural Genomics Consortium
Recommended reading
Designing an open-source microscope
Neuroscience needs a research-video archive
Explore more from The Transmitter

Building an autism research registry: Q&A with Tony Charman