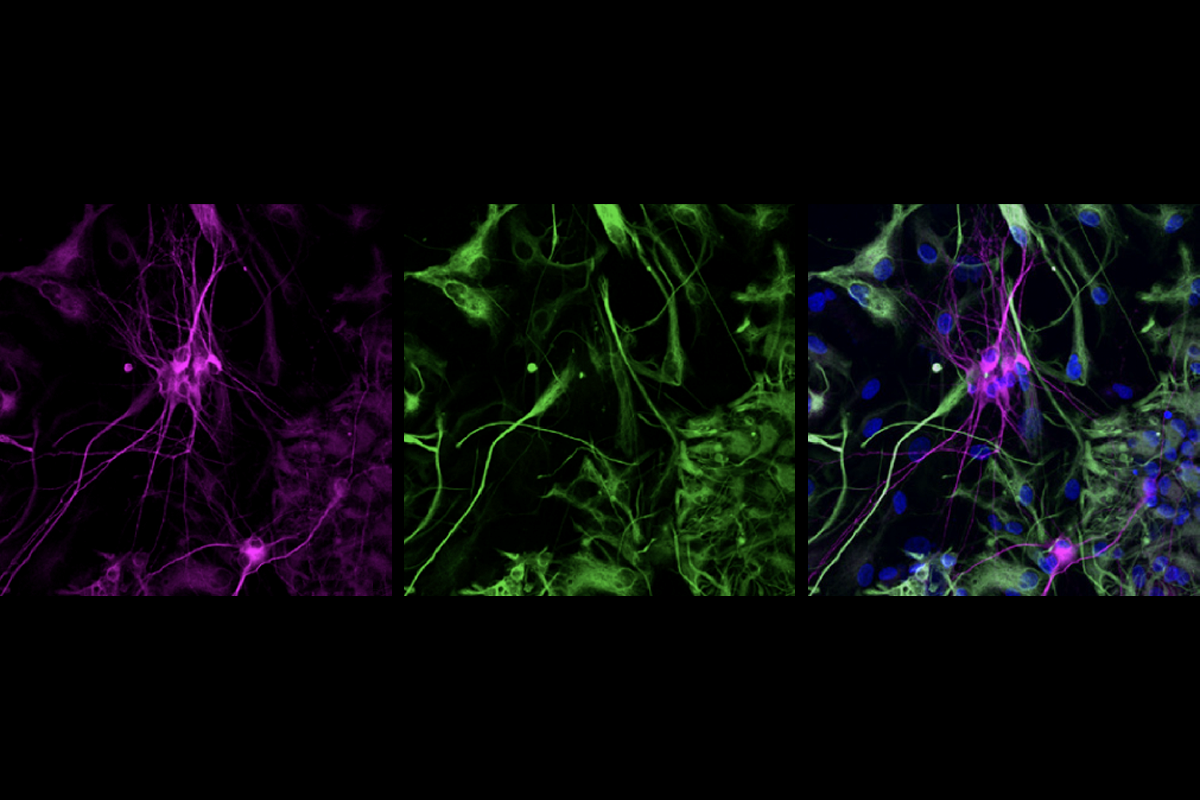
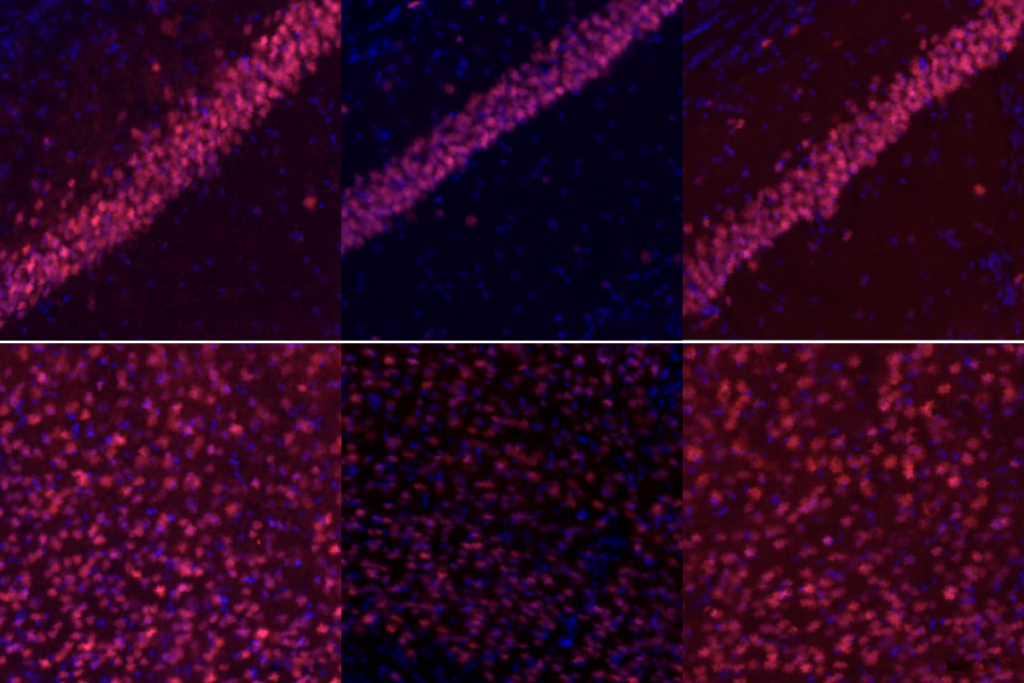
By compensating for a missing copy of SCN2A, one of the genes most strongly linked to autism, a variant of the gene-editing technology CRISPR can reduce susceptibility to seizures, according to a new study in mice.
SCN2A haploinsufficiency—in which gene variants cause someone to have just one functional copy of SCN2A instead of the usual two—occurs in about 1 in 12,000 births annually in the United States, making it one of the most prevalent genetically linked neurodevelopmental conditions.
“We want to rescue SCN2A loss,” says study investigator Kevin Bender, professor of neurology at the University of California, San Francisco. “These kids need help. This might be one avenue for treatment.”
SCN2A encodes a sodium channel that helps neurons communicate. Haploinsufficiency of the gene often leads to autism and intellectual disability, and sometimes to drug-resistant epilepsy.
Bender and his colleagues studied SCN2A haploinsufficient mice that were 30 days old, which developmentally corresponds roughly to age 10 in children. These mice have immature synapses and are more susceptible than wildtype animals to seizures induced by a chemoconvulsant.
The researchers used the technique CRISPR activation (CRISPRa) to boost expression of the intact SCN2A copy so it produced the same amount of protein as two copies of the gene. (Whereas CRISPR alters the recipient’s genome, CRISPRa enhances the expression of specific genes; Bender and his colleagues used modified CRISPR proteins to find SCN2A and bring transcriptional machinery to its promoter region.)
CRISPRa treatments delivered through an adeno-associated virus introduced either directly to the brain or intravenously to the tail restored synapse function and normalized the animals’ susceptibility to the chemoconvulsant. Bender and his colleagues detailed their findings 17 September in Nature.
“The biggest thing is not just that it worked, which is amazing, but also that it worked late in life,” Bender says.
Although the study “didn’t directly address core hallmarks of autism—for example, social impairment, repetitive behaviors, and so on—the study of SCN2A itself already makes it highly relevant to autism spectrum disorder,” says Yang Yang, associate professor of medicinal chemistry and molecular pharmacology at Purdue University, who was not involved in the work.
N
o negative side effects emerged after treatment, the researchers report. Mice with two normal copies of SCN2A that received CRISPRa began overexpressing the protein, “but that extra protein doesn’t do anything deleterious, nor does it appear to incorporate into the membrane and make cells hyper-excitable,” Bender says. The typical wildtype level of SCN2A-derived sodium channels that neurons can incorporate into their membranes may be a natural limit.In addition, the technique itself may reduce risks of gene overexpression, he and his co-authors argue. “By using CRISPRa to modify gene expression, we get to rely on the regulatory machinery of that gene normally found in cells and [that] can prove much more controlled and safe, in contrast to regular gene therapy, where you often introduce these very strong promoters and can get expression everywhere,” says study investigator Nadav Ahituv, director of the Institute for Human Genetics at the University of California, San Francisco. Regel Therapeutics, which Ahituv co-founded, has licensed this technology to treat people who have SCN2A haploinsufficiency disorders.
Still, the SCN2A haploinsufficient mice used are not good models to investigate standard autism-like behaviors, so the CRISPRa-related effects on those behaviors remain unclear, Ahituv says. The scientists are now developing a CRISPRa system for SCN2A rat models of autism, and plan to target the same promoter as in the mouse model, he adds.
A CRISPRa approach targeting enhancers instead of promoters rescues SCN2A haploinsufficiency in human cells in vitro, according to a preprint posted last year to bioRxiv.
“Both are complementary approaches,” says preprint investigator Daniel Geschwind, professor of neurology, psychiatry and biobehavioral sciences, and human genetics at the University of California, Los Angeles, who was not involved in the Nature study. “This work and ours provides strong support for this approach as a therapeutic approach that needs to be tested more broadly.”
The work is exciting for its clinical potential but needs further validation, says Karun Singh, a senior scientist at the University Health Network in Toronto, who was not involved in the preprint or the new study. That includes “testing across various patient-derived stem cell lines to determine whether the same approach can be to individuals with a different genetic background,” he says.
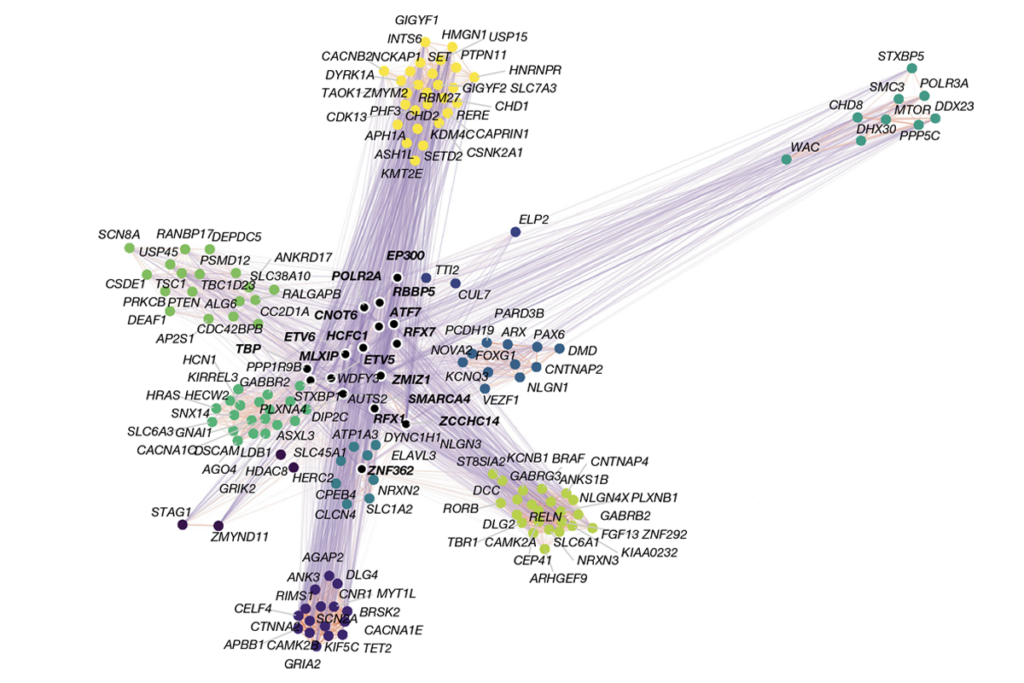
More than 200 other genes are linked to neurodevelopmental conditions due to haploinsufficiency, Bender, Ahituv and their colleagues wrote in their Nature publication. The new study could be a useful model for other work, says James Trimmer, professor emeritus of physiology and membrane biology at the University of California, Davis, who did not take part in this research. “I expect that numerous researchers will use this approach for other genes.”
The viral vectors used to deliver CRISPRa “have their limitations,” such as potential immune responses against them blunting their efficacy, Bender says. “But for better or worse, these are the same troubles that everyone in the gene therapy space is wrestling with, so we hope they can be solved, not just for this application, but for a whole slew of applications.”