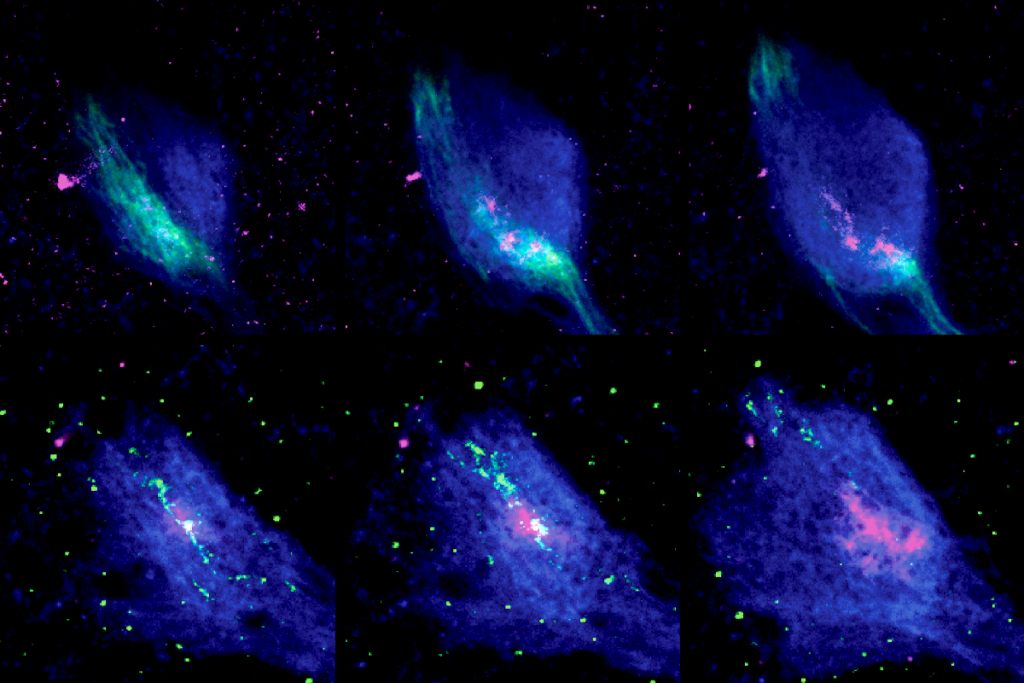
Ant olfactory neurons reveal new ‘transcriptional shield’ mechanism of gene regulation
A protective screen of spurious transcriptional activity enables each olfactory neuron to express exactly one out of hundreds of olfactory receptors.
Each of an animal’s olfactory sensory neurons expresses exactly one type of receptor encoded in its genome—a mapping that makes precise odor detection possible. Transcription factors can regulate small numbers of olfactory receptor genes; fruit flies, for example, have only 60. But that system fails in animals with many more olfactory receptors, such as ants, which rely on hundreds of rapidly evolving olfactory receptor genes to communicate with one another.
How each ant neuron expresses one and only one receptor type remained a mystery—until now: Clonal raider ants, a new study reveals, deploy a novel gene-regulation mechanism that involves a flurry of fake transcriptional activity.
“It’s a completely crazy system of transcriptional regulation—I think it’s really unprecedented,” says study investigator Daniel Kronauer, professor at Rockefeller University and a Howard Hughes Medical Institute investigator who researches social evolution and behavior in insect societies. “By studying such an extreme system with this expanded olfactory system, you can actually discover completely new mechanisms about very fundamental aspects of life.”
The olfactory receptor genes in clonal raider ants, like those in other insects, are arranged in tandem arrays, lined up one after the other in a head-to-tail fashion. Some ant arrays have upward of 80 olfactory receptor genes in a row, and the arrays appear across different chromosomes. This configuration is the result of many gene duplication events over the course of evolution, and those duplications create a thorny problem: If duplicated genes have similar sequences and machinery for regulating their expression, how do you express one receptor per neuron?
“It has been a puzzle of the highest order,” says Anandasankar Ray, professor of biology at the University of California, Riverside, who was not involved in the new work. “It’s probably developmental biology’s most complicated problem.”

W
ithin a single sensory neuron in clonal raider ants, many olfactory receptor genes are typically transcribed at once, Kronauer’s team previously found. The new work shows that when the neurons choose a transcriptional starting point among the olfactory receptor genes, that initiates a flurry of transcriptional activity—creating a kind of “transcriptional shield”—that extends more than 100 kilobases in both directions along the chromosome.In one direction, all of the genes downstream of the transcriptional starting point on the sense strand of DNA get transcribed to mRNA, resulting in dozens of transcripts of receptor genes. But only the first RNA receives a 5’ cap, a modification that allows it to be exported to the cytosol, where it is translated into protein. The other mRNA transcripts aren’t processed and never leave the nucleus, where they are eventually degraded.
In the opposite direction, polymerases transcribe RNA from the antisense strand, generating long noncoding RNAs. These transcripts effectively silence the genes on the coding strand upstream of the starting point.
After the single functional transcript is made, the remaining spurious transcriptional activity “is so massive that you can’t start canonical, real transcriptional activity anymore,” Kronauer says. It’s kind of wasteful, but it’s a “crazy and cool way” to ensure that exactly one of the many olfactory receptors gets expressed, he adds.
The mechanism has the benefit of regulating novel genes in the same way as older genes—absolving novel genes from needing to evolve new regulatory machinery, too, Kronauer adds. Whether the chromosome contains 500 or any other number of olfactory genes, the mechanism for expressing exactly one of them remains the same.
This mechanism “allows for new genes to very rapidly duplicate and integrate, and, critically, silence the other genes in the genome. So we think that this mechanism might be really important to insects with these rapidly evolving, rapidly turning-over repertoires of receptors,” says study investigator Giacomo Glotzer, a graduate student in Kronauer’s lab. The team published their findings last month in Current Biology.
The findings raise new questions, too: The researchers are investigating whether the choice of the transcriptional start site is made at random or driven by certain factors, as well as how transcriptional repression works between tandem arrays on different chromosomes. They also are working to pin down when in development the decision about which receptor to express in a given neuron is made, and how that influences subsequent development of the sensory system.
Because the specific receptor expressed by each developing sensory neuron helps determine that neuron’s wiring to the brain, the findings suggest that each ant brain is slightly different, says Christopher Potter, professor of neuroscience at Johns Hopkins University, who was not involved in the study. Each ant could express a slightly different set of odorant receptors, and the wiring of each receptor to the brain requires some flexibility. So even though clonal raider ants are all clones of one another (and therefore have identical genomes), their olfactory system may show individual variability, which could be advantageous to the colony as a whole.
Ants are an extreme case, but “my hunch is that what we discovered in the ants might be a very general phenomenon of insect receptor regulation,” Kronauer says. It’s likely to hold up in other ants and in other social insects such as honeybees, and “it might be the default mechanism. It’s just that we haven’t seen it yet, because everybody is focused on Drosophila.”
Recommended reading
Tiny eardrum sounds may help sync visual, auditory perception
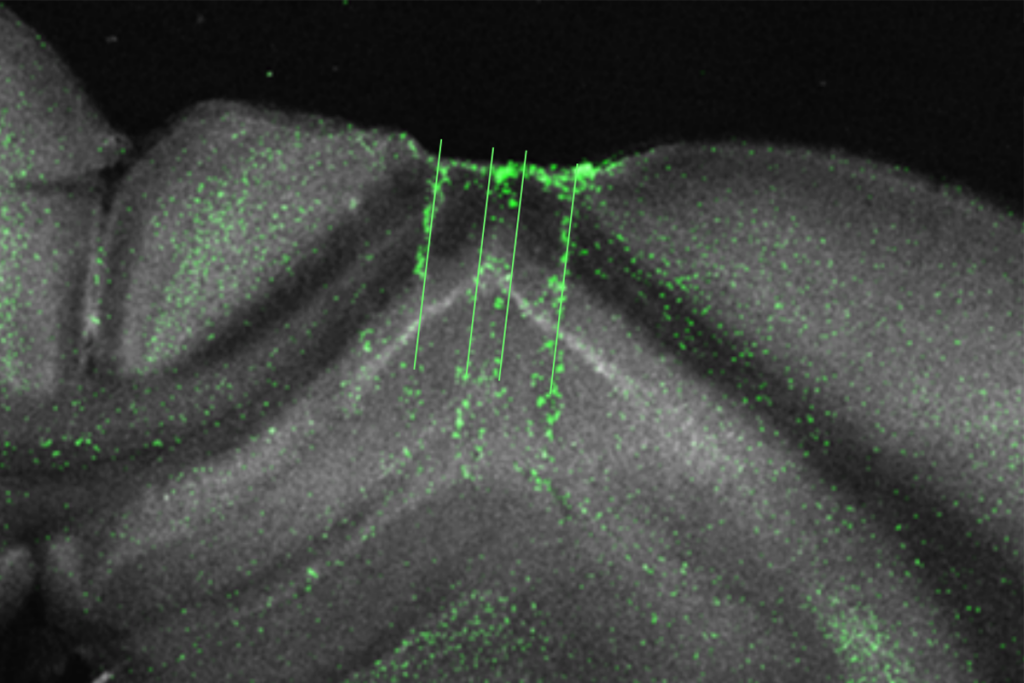
Hippocampus builds reputation as ‘general-purpose statistical learning machine’
Explore more from The Transmitter

Neuropeptides reprogram social roles in leafcutter ants
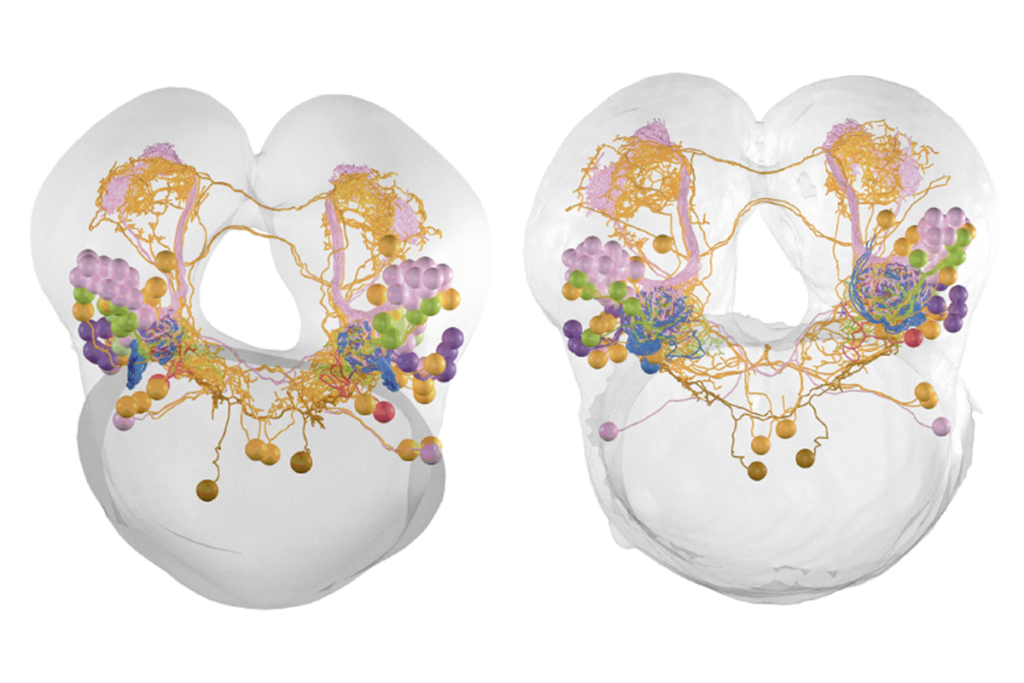
Cross-species connectome comparison shows uneven olfactory circuit evolution in flies