Molecular mechanisms: SHANK2 mutants alter synapses
Three mutations in SHANK2, an autism-associated gene, each lead to abnormal synapses, the junctions between neurons, according to a study in Human Molecular Genetics.
-
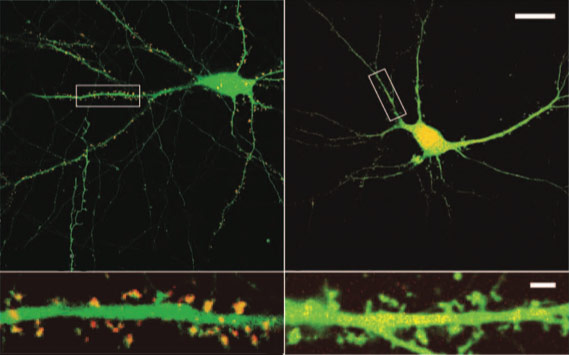
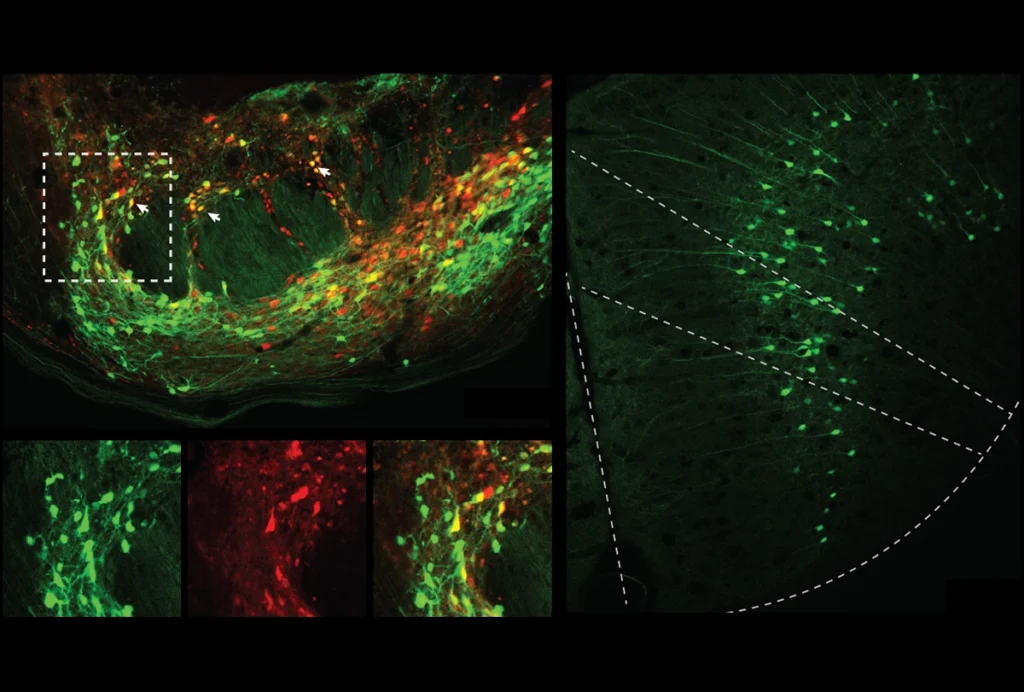
Misplaced mutant: An autism-linked variant of SHANK2 localizes throughout neurons (right) compared with the control (left), which is found only at the junctions between neurons.
- Misplaced mutant: An autism-linked variant of SHANK2 localizes throughout neurons (right) compared with the control (left), which is found only at the junctions between neurons.
Three mutations in SHANK2, an autism-associated gene, each lead to abnormal synapses, the junctions between neurons, in mice, according to a study published 27 October in Human Molecular Genetics1.
One of the three mutations, R462X, which arises de novo,meaning it is not inherited, has the most profound effect on synapses, the study found.
Over the past few years, several studies have implicated the post-synaptic density, an organized collection of proteins on the signal-receiving branches of neurons, in autism. Post-synaptic proteins linked to autism include PSD-95 and members of the SHANK protein family.
Mice lacking SHANK2 are hyperactive and have social deficits, according to unpublished data presented at the 2011 Society for Neuroscience annual meeting in Washington, D.C.
In a study published last year, researchers found several mutations in SHANK2 among individuals with autism2. In the new study, the researchers engineered mouse neurons to express one of three mutations identified, each of which affects a different region of SHANK2.
For example, L1008_P1009 is a duplication of one region of the gene, T1127M substitutes a single base pair and changes one amino acid in the protein, and R462X prematurely terminates protein expression. The duplication and the point mutant are inherited, whereas R462X is a de novo mutation.
In mouse neurons, SHANK2 localizes to dendritic spines, mushroom-shaped bodies on the signal-receiving branches of neurons. Proteins with either of the two inherited mutations localize to the same sites as the control protein does, although less of the protein is found there compared with controls, the study found.
However, R462X does not cluster at these sites, and instead is spread evenly throughout the dendrites and the cell body. Because the mutation disrupts several conserved sites, it may prevent the protein from binding to partner proteins at the post-synaptic density, the researchers say.
None of the three mutant proteins function properly at the synapse, the study found. For example, too much SHANK2 typically increases the volume of dendritic spines, but the mutant proteins do not have this effect.
Neurons lacking SHANK2 have smaller spines, fewer synapses and more complex dendritic branches compared with controls, the study found. An intact copy of SHANK2 and the L1008_P1009 duplication both rescue these effects. However, T1127M and R462X do not normalize the number of synapses, and R462X also cannot rescue the effect on dendritic branching.
The results suggest that each of the mutations has subtly different effects on the synapse, with R462X having the biggest effect.
R462X also affects neuronal signaling when injected into the mouse hippocampus, a brain region involved in learning and memory, the study found. Mice expressing R462X have less neuronal signaling in the hippocampus compared with controls.
These mice are also less likely to investigate a novel object in an open space than are controls, and are less capable at navigating a maze.
References:
1: Berkel S. et al. Hum. Mol. Genet. Epub ahead of print (2011) PubMed
2: Berkel S. et al. Nat. Genet. 42, 489-491 (2010) PubMed
Recommended reading

Building an autism research registry: Q&A with Tony Charman
CNTNAP2 variants; trait trajectories; sensory reactivity
Brain organoid size matches intensity of social problems in autistic people
Explore more from The Transmitter
Cerebellar circuit may convert expected pain relief into real thing