The spectrum goes multidimensional in search of autism subtypes
Grouping people with autism based on shared features, genetics and co-occurring conditions may improve clinical trial outcomes, researchers say.
Autism is famously varied in its presentation. All autistic people, by definition, have social communication challenges, but these can vary from mild to profound. Many have co-occurring conditions, such as anxiety or attention-deficit/hyperactivity disorder (ADHD). Some have brain regions that are atypically large or small, or connections between brain regions that are atypically strong or weak. And underpinning it all, some have single-gene disruptions, whereas others have no such obvious cause.
It has been 31 years since researchers adopted the term “spectrum” to capture autism’s heterogeneity. But the idea of a single spectrum fails to capture the many dimensions of autism, says Matthew Siegel, chief of clinical enterprise in the psychiatry and behavioral sciences department at Boston Children’s Hospital.
“The term has not served us well,” Siegel says. “We all know that categoric boundaries are porous and inaccurate. But we have to try to categorize people and understand the complexity, because that’s how we make progress.”
Siegel is among a growing group of researchers embracing the idea that there are multiple “autisms” rather than a single autism, and attempting to further categorize autistic people based on shared clinical traits as well as genetic, physiological and neuroanatomical ones. Doing so, Siegel and others argue, can reveal shared biological mechanisms and add prognostic clarity for families.
It also stands to improve the outcomes of clinical trials by putting participants “in a more homogeneous category that has better odds for [their] outcome,” says Jacob Ellegood, neuroimaging scientist at Holland Bloorview Kids Rehabilitation Hospital, who has used neuroimaging approaches in pursuit of autism subgroups.
To date, researchers have been exploring subgrouping methods with genetic, neuroimaging and clinical data, as well as developing comparison groups with animal models—with somewhat limited success. The challenge is that people with autism can be grouped in many ways, and how these differently parsed groups overlap and interact may be just as important as any one type of grouping on its own.
“We need to embrace the complexity,” says Alessandro Gozzi, director of functional neuroimaging at the Istituto Italiano di Tecnologia, who has also defined broad subgroups of autistic people based on neuroimaging. “We need many of these subgrouping modalities because each explains only some of the variability.”
T
he idea of subgrouping is nothing new. Researchers do it routinely for far less heterogeneous conditions. Autism researchers have been trying to identify subgroups based on genetics using whole-exome sequencing for more than a decade and, more recently, have used whole-genome sequencing.Such efforts have revealed some useful patterns. When autism stems from variants in CHD8, for example, it also often presents along with intellectual disability, head enlargement, speech and motor delays, sleep disturbance and gastrointestinal symptoms. Variants in other autism-linked genes, such as SHANK3 and ADNP, come with their own constellations of features.
Fewer than 30 percent of autism diagnoses are linked to specific gene disruptions, though. So researchers have looked for other ways to draw rings around people on the spectrum who may share biological features. To date, these projects have identified many groupings, but not necessarily in consistent ways or with clear clinical implications.
One new approach, described last month, sorted 5,392 autistic children in the SPARK database into four subgroups by linking common clinical features and distinct genetic signatures. (SPARK is funded by the Simons Foundation, The Transmitter’s parent organization.)
Children in the largest subgroup, representing 37 percent of those studied, have social challenges and repetitive behaviors but tend to reach developmental milestones around the same time as their neurotypical peers. Many have co-occurring conditions, such as ADHD, anxiety, depression or obsessive-compulsive disorder. Children in the smallest group—about 10 percent—tend to have more profound autism features and developmental delays. They’re also more likely than children in the other groups to have rare gene variants not inherited from a parent.
These broad subgroups are not necessarily the last word in this effort; more data may help to refine the groups further, as The Transmitter reported.
Ellegood, Gozzi and others are interested in subgrouping approaches that incorporate neuroimaging because they’re noninvasive and relatively accessible, making them relatively easy to incorporate into a diagnostic work-up.
But varied methodologies in the imaging literature have raised questions. “There’s a lot of papers coming out on autism subgrouping, and they all use slightly different data and slightly different algorithms,” says Clara Pecci Terroba, a graduate student in Richard Bethlehem’s lab at the Cambridge University Department of Psychology, who investigated these discrepancies in a study published 4 June.
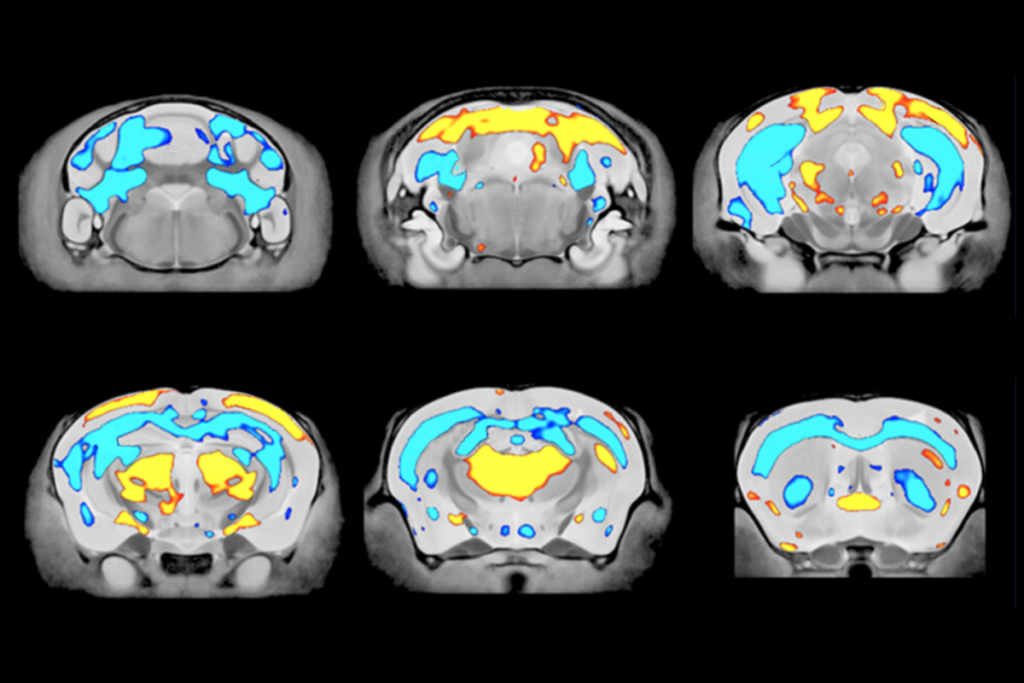
In that effort, two different machine learning algorithms applied to structural magnetic resonance imaging (MRI) data from 4,115 people—1,305 with autism, 987 with ADHD and 1,823 neurotypical controls—identified two different autism subgroups based on overall brain size. One group showed greater than typical surface area and grey matter volume and the other less; but depending on the algorithm used, some people changed groups, the researchers found. And when the team looked for differences in specific brain areas, new groups emerged.
“This tells us we should maybe spend a little bit more time trying to understand what the algorithms are doing and what they’re picking up,” Pecci Terroba says.
Even if the algorithms yielded consistent subgroupings, differences in brain surface area and grey matter volume reveal little about the mechanisms underlying autism, Siegel says. “It’s like trying to answer the question ‘What makes this car run?’ by describing the size of the car and how thick the metal is,” Siegel says. “Those are parameters, but do they really tell you anything about what makes the car go?”
S
till, identifying reproducible signals in the noise of neuroimaging data is a crucial first step, Gozzi says. Next, “we have to decode these signals into plausible, testable mechanisms.”That’s where studies that combine human and animal analyses can help, he says. For instance, four groups emerge when comparing structural MRI data from autistic people and molecular analyses from mouse models of autism, according to a preprint by Ellegood, Gozzi and their colleagues, posted on bioRxiv in March. The researchers envision being able to use the rodent model groupings as reference points for developing hypotheses about what might be happening, mechanistically, in people with parallel neurobiology but unknown genetic contributors.
And specific strategies for subtyping can help scientists bring particular research questions into focus, as a study oriented around excitation and inhibition imbalance illustrates. People with autism cluster into two groups based on electroencephalography (EEG) results—one that shows more excitation than neurotypical controls and another that shows less—according to a preprint posted to medRxiv in June by Gozzi and his colleagues. The researchers were able to recapitulate these EEG signatures by manipulating brain activity in mice, providing a model for studying how opposing patterns of brain activity can both lead to autism traits, as well as for testing different interventions.
Instead of a single spectrum, Gozzi imagines a spectrum or axis for every dimension of autism: one for core autism features, another for genetics, another for co-occurring conditions and so on. Where all of these axes intersect represents an individual’s unique “autism.”
“We started out with a very monolithic description of autism, which, clearly, is not working,” Gozzi says, adding that a description that leans heavily into individual differences may be similarly challenging for research. “We need to find some middle ground—an intermediate layer between—whereby we can find robust, reproducible groups.”
Such an approach could yield clinical progress, by identifying groups of individuals with shared features and biology who may benefit from the same treatments or supports, Gozzi says.
Siegel agrees. “It’s clear that you can’t apply the same approaches and the intervention and assistance to everybody on the spectrum,” he says, noting that the idea of autism subgroups still meets some resistance. “I think the lack of focus on, or even fear of, subgrouping—which is where we’ve been in the last 58 years—has actually caused a lot of confusion and conflation of different people’s needs.”
Recommended reading
New autism committee positions itself as science-backed alternative to government group
Astrocytes orchestrate oxytocin’s social effects in mice
Explore more from The Transmitter

Four autism subtypes map onto distinct genes, traits
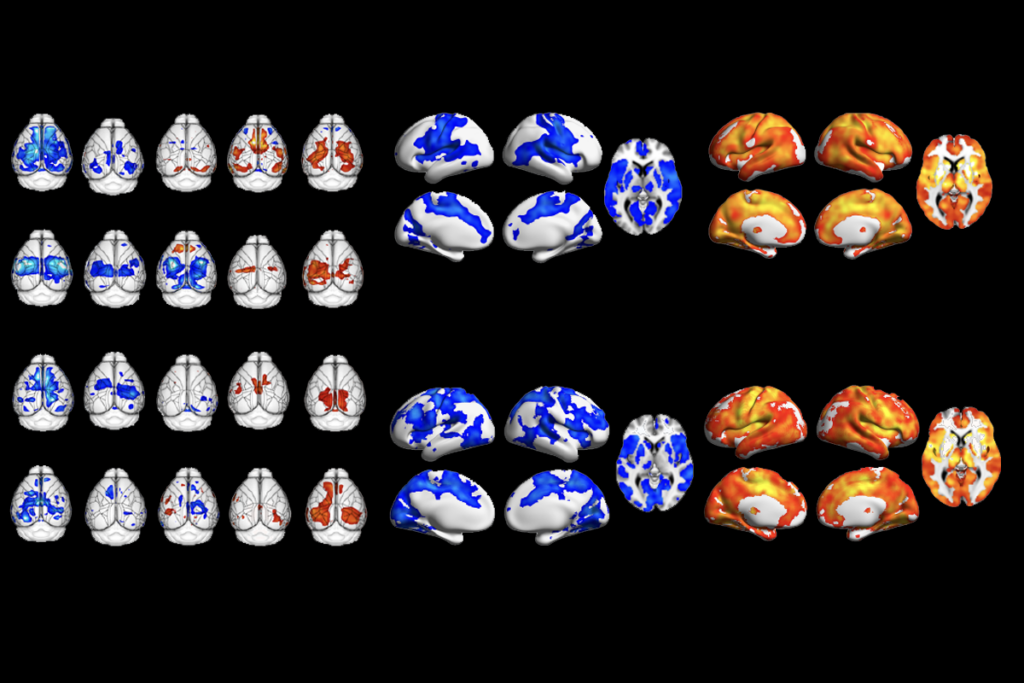
Too much or too little brain synchrony may underlie autism subtypes