One year of FlyWire: How the resource is redefining Drosophila research
We asked nine neuroscientists how they are using FlyWire data in their labs, how the connectome has transformed the field and what new tools they would like to see in the future.
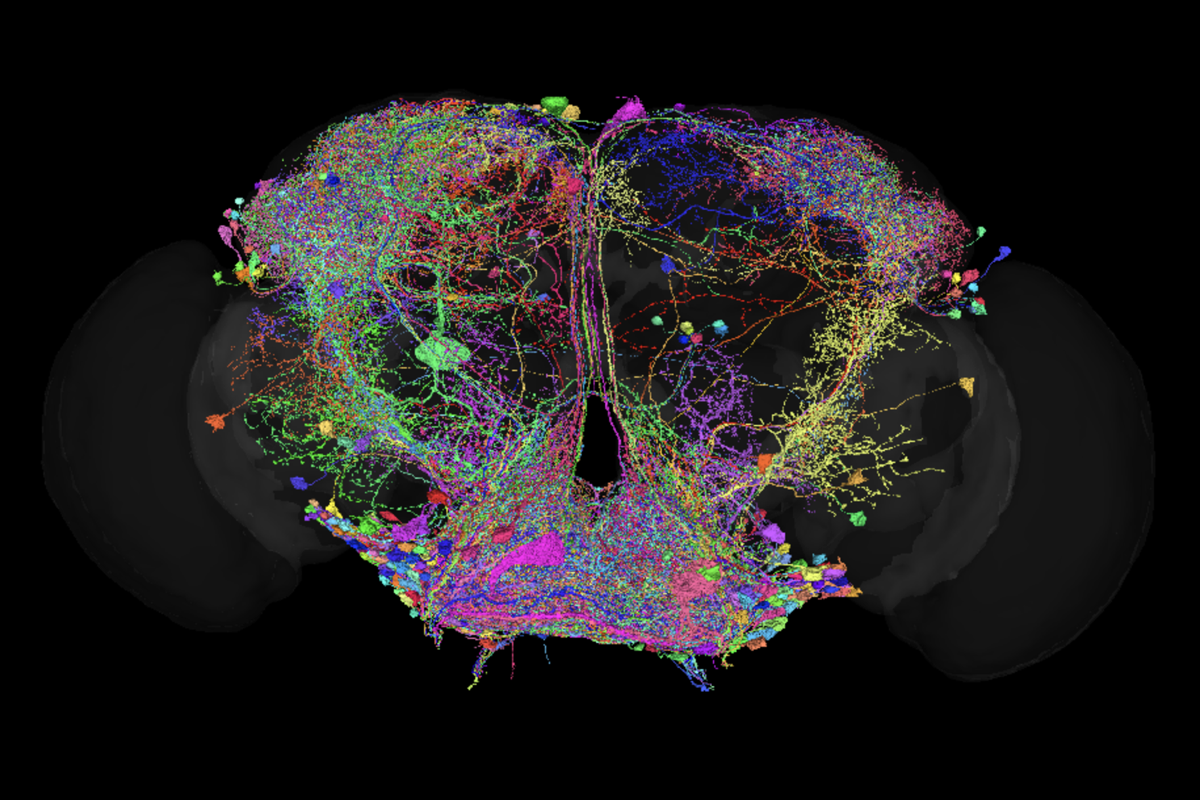
Last week marked the first anniversary of the publication of FlyWire, the first-ever complete connectome of an adult female fruit fly brain. The landmark achievement, published in a collection of papers in Nature, was the culmination of a years-long effort spearheaded by the labs of Mala Murthy and H. Sebastian Seung at Princeton University, and the FlyWire consortium, a community of neurobiologists, computer scientists and statisticians at more than 100 institutions around the world.
Mapping more than 100,000 neurons and 50 million synapses, FlyWire provides researchers with a full view of the adult female Drosophila melanogaster nervous system, dramatically speeding the pace of research. Before FlyWire, “people might spend their entire Ph.D. trying to trace the anatomy of a circuit before they could even begin to study its function,” says Anita Devineni, assistant professor of biology at Emory University. “Now we can do this with a few clicks—so we immediately know what the circuit looks like, and we can move on to the more interesting problem of studying how it works.”
Researchers have already used FlyWire data in an array of studies, describing new circuits involved in motor control and revealing novel mechanisms that regulate fly social behaviors and taste sensory processing, for example. “It’s been transformative; FlyWire has dramatically accelerated hypothesis generation and circuit mapping by making whole-brain connectivity accessible and searchable,” says Carolina Rezaval, professor of neurogenetics at the University of Birmingham. “The level of anatomical resolution and completeness also lets us think more holistically about how distributed networks coordinate behavior.”
With the connectome in hand, researchers are thinking about new tools and maps that can build on it. “I think the next big step in the connectome domain is to map the chemical connectome,” says Sung Soo Kim, assistant professor in the molecular, cellular and developmental biology department at the University of California, Santa Barbara. Other items on researchers’ wish lists include mapping connectomes from more fly species, overlaying RNA sequencing data on the connectome and integrating FlyWire with common tools, such as NeuronBridge.
To celebrate the first anniversary of FlyWire, we asked nine Drosophila researchers to share how they are using connectome data in their work, what the connectome did for the field and what new tools and resources they want for the future of Drosophila connectomics.
Responses have been lightly edited for length and clarity.
How is your lab using FlyWire connectome data in your research?
Our group is really interested in actually building these connectomes—we were one of the contributors to FlyWire. We are currently working on a male central nervous system connectome, and the FlyWire connectome has been really key in helping us work on trying to identify dimorphisms between male and female fly brains. On a general day-to-day basis, we use a lot of the annotations in the FlyWire connectome to type neurons in the male central nervous system, to identify differences in connectivity within both datasets and to look at what the consequences of these sex-specific and dimorphic neurons might be.
What new tools, analyses or additional maps would you want to see in the future?
Two things right now. We need technical advances to speed up proofreading. That is really still the bottleneck of producing connectomes. Once that becomes a little bit quicker, then creating full or partial connectomes becomes much more achievable for research groups that are not dedicated to generating connectomes. Partial connectomes could be of fly brains that are behaviorally trained and tested in the lab. A partial connectome would be restricted to a particular region of the brain, which would allow a direct link between the observed behavior and the connectivity of that individual fly. Reducing proofreading efforts will be essential for that. Having a full female central nervous system is also important. Right now, we have a female ventral nerve cord connectome, which is only partially proofread. We’re lacking a fully proofread female ventral nerve cord that we can compare with the male and get a full picture of dimorphisms between the sexes in the central nervous system.
How is your lab using FlyWire connectome data in your research?
FlyWire has been invaluable to our research. In the Drosophila taste field, the sensory neurons in the taste organs that detect different tastes are well known, but the neural circuits in the brain that receive and process taste information remained a mystery for many years—until the FlyWire connectome. As soon as the connectome came out, my lab (and others) started using it to trace the flow of taste information in the brain. With a few lines of code, we could finally identify taste pathways in the brain that our field has spent decades trying to find. My lab is now performing experiments to characterize the function of neurons in those taste pathways, and the connectome provides predictions for which stimuli each neuron responds to and what behavioral function it might have.
Every time we have a hypothesis (for example, “maybe this neuron we’re studying is involved in learning and memory”), we immediately go to FlyWire to see whether there are connections that would support our hypothesis. We still have to do the experiments, but the connectome helps us determine which experiments to do.
What did FlyWire do for the field of Drosophila neuroscience?
The FlyWire connectome has revolutionized Drosophila neuroscience. I like to say the connectome is like a road map. To get somewhere, you still need to get in the car and drive, but a road map points you in the right direction. Until now, we’ve been driving while mapping out the roads as we go, one block at a time, with only a limited understanding of the bigger picture. The connectome shows us the entire map, and with it, we might see patterns or connections that weren’t apparent with a partial view.
What new tools, analyses or additional maps would you want to see in the future?
I wanted to see a connectome that connects the brain to the nerve cord and the body (e.g., sensory organs and muscles), but thanks to a newer connectome effort, we already have this!
I would like to see additional connectomes so that we can get a better idea of how much connectivity varies by individual. In that vein, it would also be interesting to see how connectomes vary depending on the fly’s sex, age or life history. These comparisons would reveal how flexible brain connectivity is and whether some circuits are more plastic than others.
What impact has FlyWire had on your research?
FlyWire actually inspired a new collaboration. A colleague of mine, Eugenia Chiappe at the Champalimaud Centre for the Unknown, reached out to me because her lab had been doing reconstructions with FlyWire and identified some neurons that got input from halteres, a small structure in flies that plays a crucial role in flight control, which my lab currently works on. We’ve known each other for a while and had kind of been waiting for a reason to work together, and that provided the perfect opportunity. That’s the thing that’s been really exciting about FlyWire. It’s not just a tool that helps you ground your hypotheses concretely, but also by puttering around the connectome, you notice things that make you say, “Oh, I know this person who works on this.” So, I can reach out to them and chat with them about it, and then that starts a brand-new collaboration.
What did FlyWire do for the field of Drosophila neuroscience?
The level of resolution provided by FlyWire revealed connections between neurons and regions of the brain we just weren’t expecting before, which is exciting. But I think beyond the field of Drosophila neuroscience, FlyWire is a really important scaffold for neuroscience in general. This resolution of neuronal connectivity enables us to make really detailed computational models. With the help of connectome data, we can now see where models break down, which leads to questions about what that means about how a circuit is operating. Or you can see things where the model may succeed and the organism fails. FlyWire provides a really powerful way of thinking about neural circuit function. Its impact on Drosophila neuroscience is huge, but I think it has also had a big influence on how we approach doing neuroscience research in general.
What new tools, analyses or additional maps would you want to see in the future?
So, we have a few connectomes for Drosophila, and there are things that are missing in it that we’d like to have, such as electrical synapses. But I also think that we don’t need 70 Drosophila connectomes. As people get more and more interested in other animals, like mosquitoes, maybe we’d benefit from having connectomes of other related organisms. As we’ve learned from some work that came out a couple of years ago, the olfactory system in mosquitoes is very different from that of flies. What if we could compare the connectivity of a fruit fly with that of a mosquito, a hoverfly or a blowfly? This would enable us to ask questions at an evolutionary scale and investigate what kinds of things are conserved and what changes, in terms of the neural circuit function and architecture in these different animals. Thinking about how sensory information is organized centrally and how it differs in different fly families is really interesting.
How is your lab using FlyWire connectome data in your research?
We are mostly interested in comparing connectomes. Currently, we’re working on a whole new dataset: the nerve cord of Drosophila yakuba, a sister species to melanogaster. Because we are working on new datasets, we need reference connectomes like FlyWire and others to match neurons from one sister species to the other. We look at neuron morphology and how neurons connect, and then compare connectivity in different datasets to look for species differences.
How did FlyWire change Drosophila neuroscience research?
I think before the connectome, you would kind of shoot into the dark, right? Researchers would use genetically modified Gal4 flies—genetic driver lines that we use to manipulate and study neuronal activity—to activate or inhibit neurons and then see what the fly does or doesn’t do. Researchers would have to use large screens of these genetic driver lines to find the neurons that could be involved in behavior. Now, with the connectome, we have limited the neurons we need to test for because we know how they are connected. If you have an idea of one neuron that does something, and you want to find more neurons that are involved in this behavior, you can just go from the connectome and then find your genetic driver lines and just scan those. It’s helped cut down massive amounts of time and use of resources in the lab.
What impact has fly connectomics data had on the field and on your work?
First, the fly connectome data has dramatically facilitated the process of discovering the circuit mechanisms underlying specific computations. A prominent example is the description of circuits in the central complex that give rise to the representation of the animal’s head direction. Theoretical studies had long predicted that a ring attractor network can generate a unique representation of head direction, but the researchers were able to identify the exact network in the central complex using the connectome data. Second, the connectome enables researchers to generate hypotheses that lead to new insights about the function of neurons in a circuit. In a new study, we found that the output of local inhibitory neurons was critical for the detection of positive odors in a high-order area of the Drosophila olfactory system. Because there are no salient structures such as layers, columns, compartments or glomeruli in this area, without the connectome data we could not have found this local connectivity motif.
What new tools, analyses or additional maps would you want to see in the future?
The field has so far focused on analyzing the structural features of the network, but we need to understand how activity is processed in the network, which requires large-scale neural activity recording and simulation. I would therefore like to see not just a static but also a dynamic map of circuits in the future.
How is your lab using FlyWire connectome data in your research?
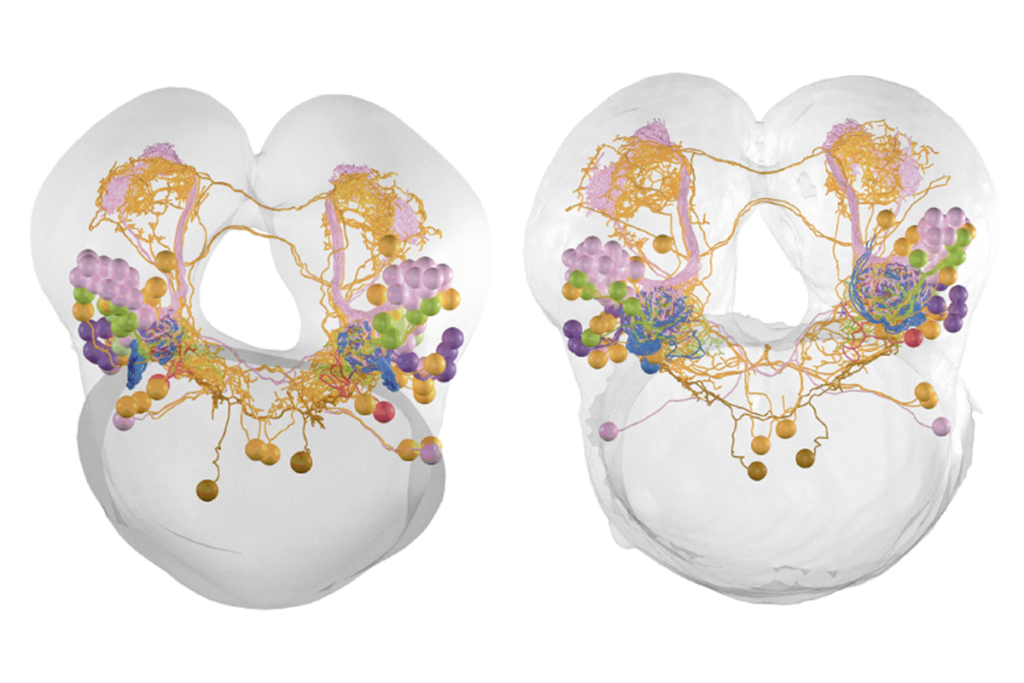
We are interested in the interface between sensory processing and cognition. Specifically, we are trying to understand how visual information is transformed into the fly’s sense of direction during flight. My lab has been using FlyWire since its early days in 2019. We’ve used the data to reconstruct the visual pathway to the central complex, a set of neuropils that play a role in sensory and motor control. We mapped the neurons from the optic lobe to the central complex, and we are using this to study the neural response of ring neurons. These neurons are connected to compass neurons that encode the sense of direction in flies. We wanted to know what kind of visual features these ring neurons encode. With this connectivity map, we can predict the functions of ring neurons, and we are now using two-photon calcium imaging and other computational modeling to test these predictions.

Courtesy of Benjamin Gorko
What new tools, analyses or additional maps would you want to see in the future?
The EM-based connectome data is wonderful. With the EM data, you can check whether a neuronal connection is real or not, just by looking at the anatomical data. There’s no ambiguity there. But the challenge right now is to figure out what those neurons are actually releasing. There are some prediction algorithms for neurotransmitter release, but so far they are not always accurate. These prediction algorithms are useful starting points. They can predict if a neuron is glutamatergic or cholinergic and so on. But if the neuron’s main releasing chemical is a neuropeptide, the only way to test for that is to actually do single-cell RNA-seq, a challenging technique that takes a lot of time. I think the next big step in the connectome domain is to map the chemical connectome. That would be awesome.
How is your lab using FlyWire connectome data in your research?
Our lab investigates how animals prioritize behaviors when confronted with competing motivations, such as whether to escape danger or secure food, and how these decisions are influenced by internal states, prior experiences and social context. A central focus of our work is understanding the role of dopamine in modulating risk-taking, motivation and behavioral flexibility. In our recent Nature paper, we found that during courtship, male flies gradually build up dopamine activity, which helps them ignore visual threats and stay focused on mating. Although dopamine is often linked to reward anticipation in mammals, we discovered that in flies it acts like a sensory filter, blocking out distractions to keep the animal focused on its goal. Using FlyWire and neuPrint, we traced the visual-threat pathway and identified downstream neurons that could be modulated by dopamine during this trade-off. Going forward, we’re using connectome data to identify novel integrator neurons that receive both sensory and neuromodulatory input. This will serve as a basis to elucidate how social context alters activity in these nodes.
What new tools, analyses or additional maps would you want to see in the future?
New tools for visually estimating synaptic weights, receptor expression data layered onto FlyWire or dynamic simulations of circuit activity would be game-changing. I’d also love to see comparative maps across developmental stages or behavioral states, which would shed light on how connectivity and function co-evolve during learning or social experience. We also need greater standardization across databases, including uniform neuron identification, to make it easier to identify genetic drivers for targeting specific neurons. Finally, we would benefit from more advanced training workshops focused on using FlyWire, especially for motif search, custom queries and integrating molecular or functional data.
How is your lab using FlyWire connectome data in your research?
In my lab, we are interested in the genetics and the molecular mechanisms of addiction. I’d say I personally go to FlyWire probably two or three times a week. If we find specific neurons that are involved in a behavior we’re studying, we want to know their input, output and their most likely neurotransmitter. As an addiction person, I’m interested in the question of motivation. We have a project going on in the lab where we have a conflict setup, where you give the flies a choice between a low-work, low-reward versus a high-work, high-reward option. The question we are interested in is whether we can overlay these behavioral decisions with some catecholaminergic modulation of the brain. We use FlyWire data to help us formulate hypotheses for this project.
What new tools, analyses or additional maps would you want to see in the future?
I think we need better integration of the connectome with other tools that are out there. Let’s say you have a neuron that is predicted to affect your behavior, and you want to manipulate it. You go to a database like NeuronBridge, which is a repository of pre-existing staining of different Gal4 lines, and you pick out a line and go perform your experiments. Integrating tools like NeuronBridge and FlyWire would make it easier for researchers to figure out how their selected Gal4 line relates to the connectome.
How is your lab using FlyWire connectome data in your research?
My lab studies how the peripheral nervous system detects social environment, and how social experience leads to changes in gene expression that reprogram brain function and behavior. We’re particularly interested in the circuits that govern pheromone detection, and—with the help of the connectome—we’re learning how these circuits relay information about the social environment to drive transcriptional and cellular changes in the brain. In a preprint we published earlier this year, we found that social experience alters behaviors by modulating transcriptional cascades, reprogramming the expression and function of circadian genes in the brain.
I also use the connectome in my neuroscience undergraduate lab course. In the class, I teach students to use the connectome to map five different olfactory circuits—three of them attractive, two of them repulsive—and we try to analyze them at multiple levels of the pathway to search for common motifs across these circuits.
What new tools, analyses or additional maps would you want to see in the future?
We need to overlay single-cell RNA-seq data onto a fully annotated connectome. Right now, my group has single-cell RNA-seq data from neurons that express fruitless, a gene involved in courtship and sex-specific behaviors, and how gene expression within this circuit changes in response to social experience. Some neurons we identify as activated or transcriptionally altered in response to group housing are annotated in the connectome, but others are not. If all of these neurons were annotated, and we had the single-cell RNA-seq data overlaid on the connectome, we could go in and easily follow all the connected neurons and observe which neural pathways are activated based on gene expression changes within the circuit. That would be incredibly useful in mapping different circuits as ensembles responding to various social environments that drive adaptive changes in brain and behavioral responses.
How are you using FlyWire in your research?
Recommended reading

The visual system’s lingering mystery: Connecting neural activity and perception

Beyond Newtonian causation in neuroscience: Embracing complex causality
Emotion research has a communication conundrum
Explore more from The Transmitter
Local circuit loops within body control fly behavior, new ‘embodied’ connectome reveals
Worms help untangle brain structure/function mystery